KU-55933
别名: ATM Kinase Inhibitor
目录号:S1092 Purity: 99.95%
KU-55933是一种有效的,特异性ATM抑制剂,在无细胞试验中IC50/Ki为12.9 nM/2.2 nM,与DNA-PK, PI3K/PI4K, ATR和mTOR相比,对ATM具有高度选择性。KU‑55933 (ATM Kinase Inhibitor)可抑制 autophagy‑initiating kinase ULK1 的激活从而导致自噬的显著减少。
CAS: 587871-26-9
客户使用Selleck的KU-55933发表文献372篇
- Nature, 2024 629(8011):443-449
- Cell, 2024 187(20):5698-5718.e26
- Cell, 2024 187(14):3652-3670.e40
- Cell, 2023 186(3):528-542.e14
- Cancer Cell, 2022 S1535-6108(22)00312-9
- Blood, 2022 blood.2021014304
- Cancer Cell, 2021 S1535-6108(20)30658-9
- Nat Genet, 2021 10.1038/s41588-021-00893-0
- Cell, 2019 178(2):361-373
- Cancer Cell, 2019 35(5):752-766
- Cancer Discov, 2019 9(9):1306-1323
- Nature, 2018 560(7716):112-116
- Cell Res, 2018 28(7):756-770
- Nat Cell Biol, 2018 20(4):413-421
- Cancer Cell, 2017 32(3):342-359
- Blood, 2017 129(21):2882-2895
- Cell, 2016 167(6):1571-1585.e18
- Science, 2016 352(6283):353-8
- Cell Discov, 2016 2:16016
- Nat Cell Biol, 2016 18(12):1357-1366
- Nature, 2015 528(7582):422-6
- Nature, 2015 10.1038/nature14328
- Nat Cell Biol, 2015 17(8):1004-13
- Nat Cell Biol, 2015 17(11):1446-57
- Mol Cancer, 2014 13:36
- Cancer Discov, 2014 4(10):1198-213
- Nat Neurosci, 2014 17(6):813-21
- Cell, 2013 21;155(5):1088-103.
- Cancer Discov, 2012 2(11):1048-63
- Nat Commun, 2024 15(1):10719
- Nat Commun, 2024 15(1):6517
- Nat Commun, 2024 15(1):10592
- Nat Commun, 2024 15(1):2089
- Mol Cell, 2024 84(21):4059-4078.e10
- Nucleic Acids Res, 2024 gkae190
- Nucleic Acids Res, 2024 gkad1224
- Nucleic Acids Res, 2024 52(6):3011-3030
- Nucleic Acids Res, 2024 gkae807
- Cell Rep Med, 2024 5(2):101393
- Clin Cancer Res, 2024 30(1):187-197
- Redox Biol, 2024 76:103323
- PLoS Biol, 2024 22(3):e3002552
- Cancer Lett, 2024 592:216898
- Cell Death Dis, 2024 15(11):799
- Cell Rep, 2024 43(2):113779
- Elife, 2024 13RP94001
- Elife, 2024 12RP91304
- Cell Oncol (Dordr), 2024 47(1):245-258
- J Biol Chem, 2024 300(12):107922
- PLoS One, 2024 19(6):e0304914
- J Cancer Res Clin Oncol, 2024 150(3):159
- bioRxiv, 2024 2024.01.03.574002
- Cell Genom, 2024 4(2):100487
- bioRxiv, 2024 2024.06.24.600514
- Nat Struct Mol Biol, 2023 10.1038/s41594-023-00929-5
- Nat Commun, 2023 14(1):6088
- Nat Commun, 2023 14(1):8293
- Adv Sci (Weinh), 2023 10(28):e2206931
- Nucleic Acids Res, 2023 10.1093/nar/gkad973
- Nucleic Acids Res, 2023 gkad533
- Nucleic Acids Res, 2023 51(13):6770-6783
- Nucleic Acids Res, 2023 51(10):4899-4913
- Cell Death Differ, 2023 10.1038/s41418-023-01167-4
- Cell Rep, 2023 42(12):113478
- Cell Rep, 2023 42(8):112805
- Cell Rep, 2023 42(11):113377
- Cell Rep, 2023 42(10):113017
- Oncogene, 2023 42(26):2113-2125
- Elife, 2023 12e87086
- Elife, 2023 12RP86976
- J Transl Med, 2023 21(1):183
- NPJ Regen Med, 2023 8(1):20
- Cells, 2023 12(19)2399
- Front Pharmacol, 2023 14:1257410
- Int J Mol Sci, 2023 24(12)10148
- Int J Mol Sci, 2023 24(3)2315
- J Biol Chem, 2023 10.1016/j.jbc.2023.105385
- Viruses, 2023 15(6)1243
- Viruses, 2023 15(5)1112
- Toxicology, 2023 484:153397
- Aquaculture, 2023 Volume 575
- Life Sci Alliance, 2023 6(4)e202201494
- Toxicol Appl Pharmacol, 2023 477:116696
- Microbiol Spectr, 2023 11(2):e0213222
- Biomol Ther (Seoul), 2023 31(5):559-565
- bioRxiv, 2023 2023.02.22.529521
- bioRxiv, 2023 2023.10.22.563489
- bioRxiv, 2023 2023.11.20.567799
- NAR Cancer, 2023 5(1):zcac045
- Nat Commun, 2022 13(1):360
- Nat Commun, 2022 13(1):4240
- Nucleic Acids Res, 2022 gkac375
- Nucleic Acids Res, 2022 50(18):10487-10502
- Cancer Res, 2022 82(24):4571-4585.
- Cancer Res, 2022 canres.2229.2021
- Cell Rep, 2022 42(1):111909
- Cell Rep, 2022 38(2):110216
- Diabetologia, 2022 10.1007/s00125-022-05810-6
- Aging Cell, 2022 21(12):e13729
- Elife, 2022 11e70726
- EMBO Rep, 2022 23(8):e54483
- Elife, 2022 11e70518
- Elife, 2022 11e74700
- Sci Signal, 2022 15(715):eabh2290
- Oncoimmunology, 2022 11(1):2016159
- Oncogenesis, 2022 11(1):33
- Cells, 2022 11(14)2149
- iScience, 2022 25(9):104892
- Open Biol, 2022 12(10):220213
- iScience, 2022 25(7):104536
- Int J Mol Sci, 2022 23(2)633
- Int J Mol Sci, 2022 23(20)12290
- Int J Mol Sci, 2022 23(13)7051
- Int J Mol Sci, 2022 23(7)3834
- Front Cell Dev Biol, 2022 10:817831
- Cancers (Basel), 2022 14(20)4984
- Cancers (Basel), 2022 14(21)5215
- Cancers (Basel), 2022 14(6)1537
- FASEB J, 2022 36(3):e22221
- J Biol Chem, 2022 S0021-9258(22)00543-9
- Viruses, 2022 14(8)1782
- PLoS One, 2022 17(8):e0271905
- Radiat Res, 2022 198(4):336-346
- Med Oncol, 2022 39(5):50
- Sci Transl Med, 2021 13(622):eabb5445
- Nat Commun, 2021 12(1):5748
- Nat Commun, 2021 12(1):3827
- Nat Commun, 2021 12(1):4373
- Mol Cell, 2021 S1097-2765(21)00315-4
- Mol Cell, 2021 S1097-2765(20)30951-5
- Nucleic Acids Res, 2021 gkaa1290
- Nucleic Acids Res, 2021 gkab584
- Hepatology, 2021 74(4):1971-1993
- Autophagy, 2021 10.1080/15548627.2021.1936932
- Cell Death Differ, 2021 10.1038/s41418-020-00733-4
- EMBO J, 2021 e106336
- J Exp Clin Cancer Res, 2021 40(1):224
- Cancer Res, 2021 81(18):4676-4684
- Clin Transl Med, 2021 11(10):e608
- Cell Rep, 2021 34(7):108713
- Cell Rep, 2021 34(9):108808
- Cell Rep, 2021 34(7):108744
- Cell Rep, 2021 35(13):109306
- Oncogene, 2021 10.1038/s41388-021-01932-0
- Cell Mol Life Sci, 2021 10.1007/s00018-021-03902-x
- J Cell Biol, 2021 220(2)e201807163
- Sci Signal, 2021 14(705):eabc4764
- Cell Mol Gastroenterol Hepatol, 2021 S2352-345X(21)00249-6
- Commun Biol, 2021 4(1):484
- Front Cell Dev Biol, 2021 9:649328
- Front Cell Dev Biol, 2021 9:810928
- Front Cell Dev Biol, 2021 9:650819
- Front Cell Dev Biol, 2021 9:662868
- J Cell Mol Med, 2021 10.1111/jcmm.16751
- Cancers (Basel), 2021 13(15)3843
- Eur J Pharmacol, 2021 907:174303
- Carcinogenesis, 2021 bgab002
- J Oncol, 2021 2021:8239984
- DNA Repair (Amst), 2021 104:103136
- Hum Mol Genet, 2021 ddab300
- Chembiochem, 2021 22(12):2177-2181
- FEBS Open Bio, 2021 10.1002/2211-5463.13152
- Cancer Invest, 2021 1-12
- Sci Adv, 2021 7(25)eabd9208
- Methods Mol Biol, 2021 2267:103-144
- Research Square, 2021 10.21203/rs.3.rs-162979/v1
- Nat Commun, 2020 11(1):5775
- Nat Commun, 2020 11(1):6178
- Nat Commun, 2020 11(1):123
- Nat Commun, 2020 26;11(1):2641
- Nat Commun, 2020 8;11(1):2876
- Mol Cell, 2020 S1097-2765(20)30723-1
- Mol Cell, 2020 77(5):1153
- Mol Cell, 2020 22 pii: S1097-2765(20)30231-8
- Adv Sci (Weinh), 2020 7(20):2000157
- Nucleic Acids Res, 2020 6;48(6):2982-3000
- Nat Chem Biol, 2020 10.1038/s41589-020-0600-3
- Cell-Rep, 2020 18;30-7-:2402-2415-e5
- Cell Rep, 2020 18;30(7):2083-2093 e5
- Cell Rep, 2020 14;31(2):107518
- Cell Rep, 2020 32(9):108080
- Cell Rep, 2020 31(10):107745
- Arterioscler Thromb Vasc Biol, 2020 ATVBAHA120315206
- Cell Mol Biol Lett, 2020
- Cell Mol Life Sci, 2020 77(4):735-749
- Elife, 2020 30;9:e51466
- PLoS Pathog, 2020 16(6):e1008514
- Mol Oncol, 2020 26
- mBio, 2020 11(1)
- mBio, 2020 11(3):e01190-20
- Arch Toxicol, 2020 10.1007/s00204-020-02712-7
- Front Pharmacol, 2020 11:601468
- Aging (Albany NY), 2020 22;12(6):4688-4710
- Cancers (Basel), 2020 28;12(6):E1388
- Cancers (Basel), 2020 12(6):1490
- Cancers (Basel), 2020 12(9):E2356
- Cancers (Basel), 2020 12(9)E2467
- J Biol Chem, 2020 295(1):125-137
- Sci Rep, 2020 10(1):2200
- Sci Rep, 2020 10(1):14455
- DNA Repair (Amst), 2020 87:102803
- PLoS One, 2020 15(1):e0223463
- Int J Radiat Biol, 2020 96(4):461-468
- Med Mol Morphol, 2020 10.1007/s00795-020-00264-4
- Nat Commun, 2019 10(1):2910
- Nat Commun, 2019 10(1):1577
- Nat Commun, 2019 10(1):87
- Mol Cell, 2019 75(6):1299-1314
- Mol Cell, 2019 74(4):651-663.e8
- J Exp Med, 2019 216(7):1648-1663
- Nucleic Acids Res, 2019 47(10):5243-5259
- Nucleic Acids Res, 2019 47(21):10977-10993
- Nucleic Acids Res, 2019 10.1093/nar/gkz518
- Cell Death Differ, 2019 10.1038/s41418-019-0393-7
- Cancer Res, 2019 10.1158/0008-5472.CAN-18-3388
- J Pineal Res, 2019 67(4):e12603
- Haematologica, 2019 10.3324/haematol.2018.215210
- Cell Rep, 2019 26(8):2194-2211.e6
- Cell Rep, 2019 28(3):735-745
- J Cell Biol, 2019 218(7):2113-2123
- Oxid Med Cell Longev, 2019 2019:8535163
- Int J Radiat Oncol Biol Phys, 2019 105(4):861-874
- Mol Oncol, 2019 13(2):290-306
- Mol Cancer Ther, 2019 18(8):1439-1450
- J Cell Physiol, 2019 234(11):21294-21306
- J Virol, 2019 93(14)
- Cancers (Basel), 2019 11(12)
- J Biol Chem, 2019 294(17):7037-7045
- Neoplasia, 2019 21(12):1143-1150
- J Biol Chem, 2019 294(8):2827-2838
- Sci Rep, 2019 9(1):13097
- Retrovirology, 2019 16(1):13
- Sci Adv, 2019 5(3):eaav1118
- Sci Adv, 2019 5(5):eaav3235
- Nat Struct Mol Biol, 2018 25(3):261-269
- Nat Commun, 2018 9(1):697
- Nat Commun, 2018 9(1):5351
- Nat Commun, 2018 9(1):1317
- Nat Commun, 2018 13;9(1):2720
- Mol Cell, 2018 71(2):332-342
- Mol Cell, 2018 69(5):879-892.e5
- Nucleic Acids Res, 2018 46(16):8311-8325
- Nucleic Acids Res, 2018 46(19):10119-10131
- Redox Biol, 2018 14:645-655
- Cancer Res, 2018 78(18):5363-5374
- Cancer Lett, 2018 419:280-290
- Cell Death Dis, 2018 9(2):164
- Cell Death Dis, 2018 9(6):660
- Cell Death Dis, 2018 9(2):146
- Cell Rep, 2018 24(13):3393-3403.e5
- Acta Pharmacol Sin, 2018 39(10):1645-1660
- Acta Pharmacol Sin, 2018 39(12):1874-1884
- Int J Cancer, 2018
- Mol Cancer Ther, 2018 17(12):2551-2563
- Am J Cancer Res, 2018 8(9):1697-1711
- Int J Oncol, 2018 52(3):709-720
- Transl Oncol, 2018 11(4):988-998
- FASEB J, 2018 32(4):1818-1829
- Oncol Rep, 2018 39(5):2071-2080
- Oncol Rep, 2018 40(4):2363-2370
- Arch Biochem Biophys, 2018 644:93-99
- Virology, 2018 523:121-128
- Hum Mol Genet, 2018 27(23):4024-4035
- Radiat Res, 2018 189(5):505-518
- Oncol Lett, 2018 16(5):6608-6614
- Sci Adv, 2018 4(12):eaat5077
- Radiat Environ Biophys, 2018 57(1):31-40
- Nat Commun, 2017 8(1):2039
- Nucleic Acids Res, 2017 45(18):10564-10582
- Genome Biol, 2017 18(1):224
- EMBO J, 2017 2;36(9):1261-1278
- J Exp Clin Cancer Res, 2017 36(1):133
- Cancer Res, 2017 77(17):4567-4578
- Genes Dev, 2017 31(7):648-659
- Genes Dev, 2017 31(3):318-332
- Sci Total Environ, 2017 1599:375-390
- Cancer Lett, 2017 390:48-57
- Cell Death Dis, 2017 8(8):e3019
- Cell Death Dis, 2017
- J Cell Biol, 2017 216(3):623-639
- EMBO Rep, 2017 18(5):781-796
- Free Radic Biol Med, 2017 108:750-759
- Stem Cells Transl Med, 2017 6(6):1504-1514
- Am J Physiol Cell Physiol, 2017 313(5):C501-C515
- Int J Oncol, 2017 50(1):290-296
- J Biol Chem, 2017 292(30):12424-12435
- Sci Rep, 2017 7(1):16115
- Sci Rep, 2017 7(1):5432
- Eur J Pharm Sci, 2017 109:223-232
- Oncol Rep, 2017 38(1):591-597
- BMC Cancer, 2017 17(1):338
- Virology, 2017 510:9-21
- PLoS One, 2017 12(7):e0181530
- Nutr Cancer, 2017 69(8):1281-1289
- Mol Cell, 2016 61(3):405-418
- Nucleic Acids Res, 2016 44(10):4745-62
- Genes Dev, 2016 30(3):321-36
- Oncogene, 2016 35(5):567-76
- J Cell Biol, 2016 212(4):399-408
- Elife, 2016 10.7554/eLife.15129
- Oxid Med Cell Longev, 2016 2016:4313204
- Cell Death Discov, 2016 2:16011
- Oncotarget, 2016 7(38):60807-60822
- Oncotarget, 2016 7(20):29359-70
- Oncotarget, 2016 7(31):49800-49818
- FASEB J, 2016 30(8):2899-914
- Sci Rep, 2016 6:22972
- Sci Rep, 2016 6:37194
- Sci Rep, 2016 6:27379
- Scientific Reports, 2016 37194-
- J Immunol, 2016 197(7):2918-29
- Cell Cycle, 2016 15(1):84-94
- Chem Res Toxicol, 2016 29(1):26-39
- J Cell Sci, 2016 129(16):3167-77
- Toxicol Appl Pharmacol, 2016 292:65-74
- BMC Cancer, 2016 16(1):725
- PLoS One, 2016 11(12):e0167454
- Cancer Chemother Pharmacol, 2016 77(1):169-80
- Mutat Res Genet Toxicol Environ Mutagen, 2016 795:1-6
- Leukemia, 2015 29(5):1133-42
- Genes Dev, 2015 29(11):1151-63
- Oncogene, 2015 34(23):2978-90
- Mol Cell Proteomics, 2015 14(8):2261-73
- Nanoscale, 2015 7(47):20211-9
- Arch Toxicol, 2015 89(11):2015-25
- Am J Cancer Res, 2015 5(5):1649-64
- Mol Cancer Res, 2015 13(1):120-9
- Cell Signal, 2015 27(5):951-60
- J Biol Chem, 2015 290(46):27545-56
- Sci Rep, 2015 5:11465
- PLoS Genet, 2015 11(10):e1005563
- Cell Cycle, 2015 14(20):3248-60
- Cell Cycle, 2015 14(4):648-55
- Cell Biol Int, 2015 10.1002/cbin.10439
- PLoS One, 2015 10(11):e0142561
- PLoS One, 2015 10(12):e0145994
- Cancer Biol Ther, 2015 16(11):1585-92
- Mol Biol Cell, 2015 26(12):2217-26
- Int J Radiat Biol, 2015 10.3109/09553002.2015.1029595
- Anticancer Res, 2015 35(7):3829-38
- Cell Death Differ, 2014 10.1038/cdd.2014.159
- Oncogene, 2014 10.1038/onc.2014.268
- J Biol Chem, 2014 289(23):16072-84
- Biochim Biophys Acta, 2014 1843(5):934-44
- Biochim Biophys Acta, 2014 1842(7):1071-9
- Biochim Biophys Acta, 2014 1842(7):1088-96
- Cell Cycle, 2014 13(4):666-77
- Cell Cycle, 2014 13(11):1777-87
- DNA Repair, 2014 21:140-7
- Toxicol Appl Pharm, 2014 282(2):227-236
- PLoS One, 2014 9(6):e98891
- Radiat Res, 2014 181(6):650-8
- Proteomes, 2014 2(3):451-467
- Theriogenology, 2014 12(5):e1001856
- Julius Maximilians Universit, 2014 Kumari G
- Nucleic Acids Res, 2013 41(22):10157-69
- Sci Signal, 2013 6(259):ra5
- Cell Cycle, 2013 12(7):1105-18
- J Gen Virol, 2013 94(Pt 12):2664-2669
- Neurotox Res, 2013 25(2):193-207
- PLoS One, 2013 9(1):e84899
- PLoS One, 2013 8(11):e80313
- FEBS Lett, 2013 587(21):3437-43
- Autophagy, 2012 8(2):236-51
- J Biol Chem, 2012 287(26):22112-22
- J Biol Chem, 2012 287(12):8803-15
- J Immunol, 2012 188(5):2266-75
- Food Chem Toxicol, 2012 55:214-21
- Int J Radiat Biol, 2012 88(7):507-14
- J Virol, 2011 85(23):12547-56
- Mech Ageing Dev, 2011 132(11-12):543-51
- J Biol Chem, 2011 286(10):8394-404
化学信息&溶解度
| 分子量 | 395.49 |
| 分子式 | C21H17NO3S2 |
| CAS号 | 587871-26-9 |
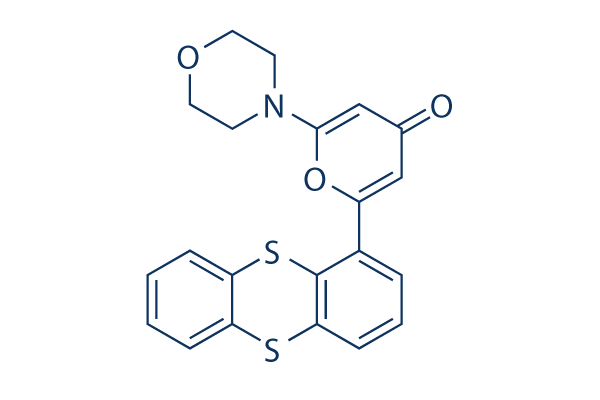
| Smiles | C1COCCN1C2=CC(=O)C=C(O2)C3=C4C(=CC=C3)SC5=CC=CC=C5S4 |
| 储存条件(自收到货起) | |
建议分装储备液,避免反复冻融! |
|
| 体外溶解度 | 批次: |
|
DMSO : 39 mg/mL ( 98.61 mM; DMSO吸湿会降低化合物溶解度,请使用新开封DMSO) Water : Insoluble Ethanol : Insoluble DMSO : 79 mg/mL ( 199.75 mM; DMSO吸湿会降低化合物溶解度,请使用新开封DMSO) Water : Insoluble Ethanol : Insoluble DMSO : 33 mg/mL ( 83.44 mM; DMSO吸湿会降低化合物溶解度,请使用新开封DMSO) Water : Insoluble Ethanol : Insoluble DMSO : 79 mg/mL ( 199.75 mM; DMSO吸湿会降低化合物溶解度,请使用新开封DMSO) Water : Insoluble Ethanol : Insoluble |
|
| 体内溶解度 | 批次: |
| 现配现用,请按从左到右的顺序依次添加,澄清后再加入下一溶剂 | |
|
5%DMSO
Corn oil
浓度:2mg/ml
(5.06mM)
操作示例:以 1 mL 工作液为例,取50μL40mg/ml的澄清DMSO储备液加到950μL玉米油中,混合均匀。工作液请现配现用!
|
|
|
5%DMSO
40%PEG300
5%Tween80
50%ddH2O
浓度:3.95mg/ml
(9.99mM)
操作示例:以 1 mL 工作液为例,取50μL79mg/ml的澄清DMSO储备液加到 400μL PEG300中,混合均匀使其澄清;向上述体系中加入50μLTween80,混合均匀使其澄清;然后继续加入500μL ddH2O定容至 1 mL。工作液请现配现用!
|
|
摩尔浓度计算器
质量(g)= 浓度(mol/L)* 体积(L)* 分子量(g/mol)
动物体内配方计算器(澄清溶液)
第一步:请输入基本实验信息(考虑到实验过程中的损耗,建议多配一只动物的药量)
第二步:请输入动物体内配方组成(配方适用于不溶于水的药物;不同批次药物配方比例不同,请联系Selleck为您提供正确的澄清溶液配方)
计算结果:
工作液浓度: mg/ml;
DMSO母液配制方法: mg 药物溶于 μL DMSO溶液(母液浓度 mg/mL,注:如该浓度超过该批次药物DMSO溶解度,请先联系Selleck);
体内配方配制方法:取 μL DMSO母液,加入 μL PEG300,混匀澄清后加入 μL Tween 80,混匀澄清后加入 μL ddH2O,混匀澄清。
体内配方配制方法:取 μL DMSO母液,加入 μL Corn oil,混匀澄清。
注意:
1. 首先保证母液是澄清的;
2.一定要按照顺序依次将溶剂加入,进行下一步操作之前必须保证上一步操作得到的是澄清的溶液,可采用涡旋、超声或水浴加热等物理方法助溶。

